Towards safer and efficient formulations: Machine learning approaches to predict drug-excipient compatibility
Abstract
Predicting drug-excipient compatibility is a critical aspect of pharmaceutical formulation design. In this study, we introduced an innovative approach that leverages machine learning techniques to improve the accuracy of drug-excipient compatibility predictions. Mol2vec and 2D molecular descriptors combined with the stacking technique were used to improve the performance of the model. This approach achieved a significant advancement in the predictive capacity as demonstrated by the accuracy, precision, recall, AUC, and MCC of 0.98, 0.87, 0.88, 0.93 and 0.86, respectively. Using the DE-INTERACT model as the benchmark, our stacking model could remarkably detect drug-excipient incompatibility in 10/12 tested cases, while DE-INTERACT managed to recognize only 3 out of 12 incompatibility cases in the validation experiments. To ensure user accessibility, the trained model was deployed to a user-friendly web platform. This interactive interface accommodated inputs through various types, including names, PubChem CID, or SMILES strings. It promptly generated compatibility predictions alongside corresponding probability scores. However, the continual refinement of model performance is crucial before applying this model in practice.
Introduction
Excipients are inactive compounds incorporated into the formulation to enhance the stability and bioavailability of active pharmaceutical ingredients (APIs) (Jackson et al., 2000). While generally considered as inert, excipients can, in specific cases, initiate undesirable interactions with active substances, subsequently undermining the bioavailability of APIs (Sims et al., 2003). This underscores the significance of the investigation of drug-excipient compatibility, which significantly impacts the optimization of drug formulation and the development of drug delivery systems.
To explore the compatibility between drugs and excipients, a commonly used approach involves mixing them at precise ratios and storing the mixtures at specific conditions. Any resulting interactions are then identified using various techniques, such as High-Performance Thin Layer Chromatography (HPTLC), High-Performance Liquid Chromatography (HPLC), Differential Scanning Calorimetry (DSC), and Infrared Spectra (IR spectra) (Chadha and Bhandari, 2014). Despite their precision in detecting drug-excipient incompatibility, these methods frequently require significant time and sophisticated instrumentation. As a result, it is necessary to find an alternative strategy that can provide rapid and accurate results. For example, Defang Ouyang, et al., utilized a knowledge-driven expert system called PharmDE, which demonstrated highly encouraging successes in identifying possible incompatibilities that may occur between active pharmaceutical ingredients and excipients (Wang et al., 2021).
Machine learning, a subset of AI, is playing an increasingly pivotal role in the realm of drug discovery and development. Its capacity to handle large datasets and discern intricate patterns offers the potential to accelerate the drug discovery process, ultimately enhancing both drug efficacy and safety (Vamathevan et al., 2019). Currently, machine learning finds application in tasks such as identifying potential drug targets (Mayr et al., 2018), forecasting drug efficacy and toxicity (Vo et al., 2019), and optimizing formulations and dosing regimens (Bannigan et al., 2021). In 2023, S. Patel, et al., harnessed the power of machine learning to predict the interactions between small-molecule drugs and excipients, using 881-bit binary fingerprints of each drug and excipient as molecular descriptors. The results of their study revealed that the developed model, known as DE-INTERACT, exhibited remarkable accuracy and the ability to rapidly predict instances of drug-excipient incompatibility (Patel et al., 2023).
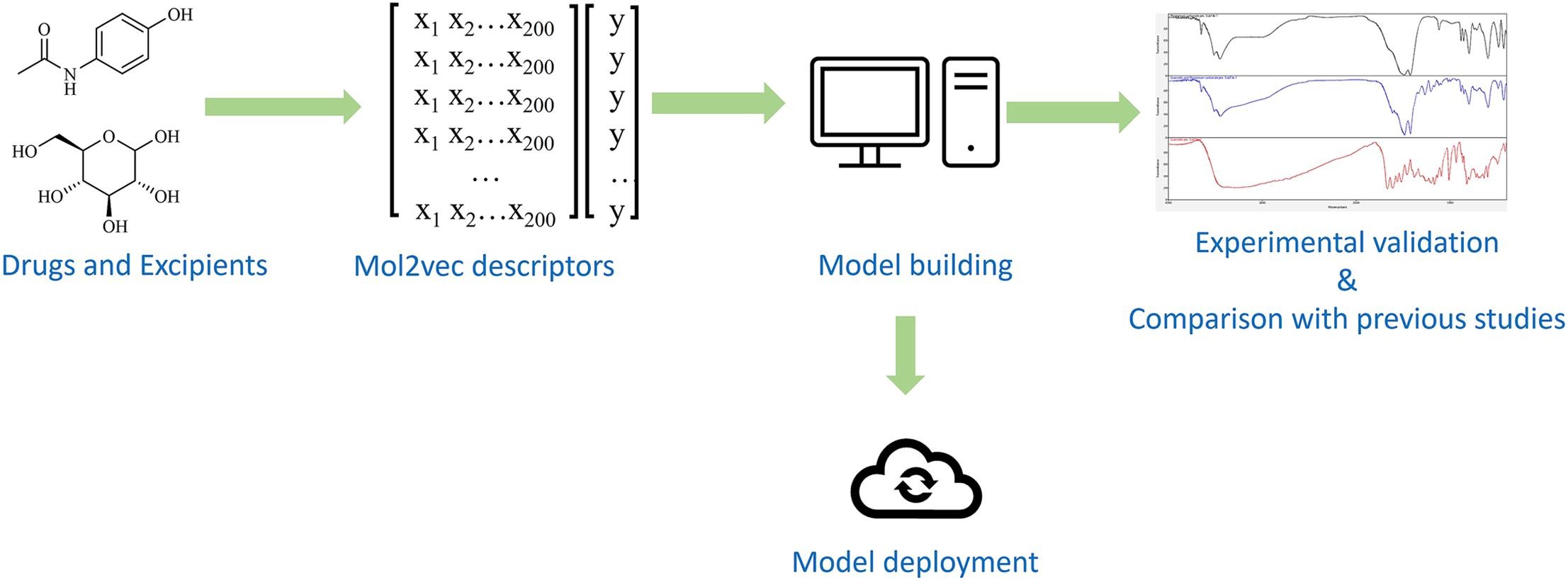
Natural language processing represents a crucial branch of machine learning with wide-ranging implications across diverse fields, including chemistry (Hirschberg and Manning, 2015). Drawing inspiration from the word2vec model, a prevalent technique used in natural language processing, S. Jaeger, et al. introduced the mol2vec method. This innovative approach involved transforming a chemical molecule into a vector, which could be used as inputs for machine learning algorithms (Jaeger et al., 2018). Numerous studies have highlighted the effectiveness of this method, often yielding outcomes on par with or even surpassing conventional molecular descriptors (Jaeger et al., 2018, Parakkal et al., 2022, Sato et al., 2022, Shibayama et al., 2020).
Therefore, our study aimed to construct a robust machine learning model capable of accurately predicting the compatibility between drugs and excipients within a formulation, utilizing the mol2vec technique. To evaluate its performance, the model’s results were compared with those of a previous study conducted by S. Patel, et al. The accuracy and reliability of our developed model were also substantiated through experimental validation.
Read more here
Nguyen Thu Hang, Nguyen Thanh Long, Nguyen Dang Duy, Nguyen Ngoc Chien, Nguyen Van Phuong,
Towards safer and efficient formulations: Machine learning approaches to predict drug-excipient compatibility,
International Journal of Pharmaceutics, Volume 653, 2024, 123884, ISSN 0378-5173, https://doi.org/10.1016/j.ijpharm.2024.123884.
Read more articles on Machine Learning here:
- Using AI/ML to predict blending performance and process sensitivity for Continuous Direct Compression
- A dataset of formulation compositions for self-emulsifying drug delivery systems
- Data-driven development of an oral lipid-based nanoparticle formulation of a hydrophobic drug